Results of CACHE Challenge #4
CACHE#4 participants who asked to be de-anonymized:
- Keunwan Park, Charuvaka Muvva, Sriramulu Dinesh Kumar (Korea Institute of Science and Technology).
- Wei Lu, Jixian Zhang (Galixir).
- Shuangjia Zheng (Shanghai Jiaotong University).
- Sandro Cosconati, Michele Roggia (University of Campania Luigi Vanvitelli).
- Olga Tarkhanova, Mykola Protopopov, Yurii Moroz (Chemspace LLC).
- Douglas Houston, Vincent Blay-Roger, Gediminas Gumbis, Steve Shave, Yan-Kai Chen, Miles McGibbon, Yumiao Gao, Lin Chen, Boyang Ni (University of Edinburgh).
- Jude Wells, Edina Rosta, Dénes Berta, Wim Dehaen, Andrea Karlova, Sam Martino, Christine Orengo, Brooks Paige, Neeladri Sen, Geoff Wells, Sarah Harris, Daniel Felfoldi, Akshay Sethia, Alice Cheng, Ferdie Krammer, Gbetondji Dovonon, (University College London).
- Alexander Tropsha, Dima Kozakov, Konstantin Popov (University of North Carolina at Chapel Hill).
CACHE#4 compounds were tested experimentally at the Structural Genomics Consortium, University of Toronto and University Health Network with contributions from Cheryl Arrowsmith, Albina Bolotokova, Irene Chau, Kristina Edfeldt, Elisa Gibson, Levon Halabelian, Rachel Harding, Oleksandra Herasymenko, Scott Houliston, Ashley Hutchinson, Peter Loppnau, Matthieu Schapira, Almagul Seitova and Madhushika Silva, Dakota Treleaven, and Hong Wu.
SUMMARY
23 computational teams selected up to 100 compounds each. The resulting 1688 compounds were tested experimentally. After a hit expansion round, teams were ranked based on the activity and novelty of their predicted molecules. Results are summarized the Table below. One team was successful in generating a clearly novel chemical hit with strong experimental validation. A different molecule predicted by another team also showed convincing experimental confirmation but was chemically related to CBLB ligands previous disclosed in the patent literature. Other molecules were either inactive or not firmly confirmed experimentally speaking to the challenging nature of this highly dynamic target.
INTRODUCTION
- For CACHE #4, 23 participants from 12 countries used their computational workflows to predict small molecule ligands that bind to the close/inactive conformational state of the TKB domain of the E3 ligase CBLB, an immuno-oncology target. At the start of this challenge, over 800 ligands had been reported, all chemically related. A few weeks before the challenge started, a crystal structure of CBLB in complex with one of the known ligands was released (PDB 8GCY), revealing a conformational remodeling of the target and providing structural insight for the design of novel inhibitors. Given this information, the CACHE #4 challenge was to design chemically novel ligands.
- CACHE Challenges begin with a hit finding round (Round 1), where participants can nominate up to 100 commercially available compounds. Round 1 predictions are then experimentally tested and the resulting data returned to participants. In the hit-expansion round (Round 2), participants whose nominated compounds show some sign of activity in experimental binding assays (compounds of interest) can select up to 50 follow-up compounds. Together, these two rounds are designed to avoid both false positives and false negatives, which is critical to evaluate computational methods properly.
- In a separate exercise, computational teams are asked to predict hits from a library composed of the merged Round 1 selections from all participants. Here, all teams are screening the same library, and all compounds are experimentally characterized as active or inactive, but experimental data is blinded to participants.
THE CHALLENGE
- CACHE participants were asked to predict compounds binding the TKB domain of CBLB with novel chemical templates.
- After a double-blind peer review where each applicant reviewed 5 applications, 23 participants joined and completed the challenge, representing a diverse array of physics-based and AI computational methods.
- Participants collectively selected 1688 compounds (no more than 100 compounds per participant) that were ordered from Enamine and tested experimentally.
ROUND 1 RESULTS
All Round 1 experimental data are provided here and here
1688 compounds were screened in two independent runs at 50 µM in a fluorescence polarization (FP) assay which measured displacement of the fluorescently labelled version of the known CBLB ligand C2787102.
FP Assay conditions: recombinant CBLB [construct 34-427 with C-terminal Avi and N-terminal His tagged] at 625 nM; BODIPY-labeled C2787102 at 10 nM; Buffer: 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 0.01% (v/v) TritionX-100, 2% (v/v) DMSO.
- A good correlation was observed between the two independent runs:
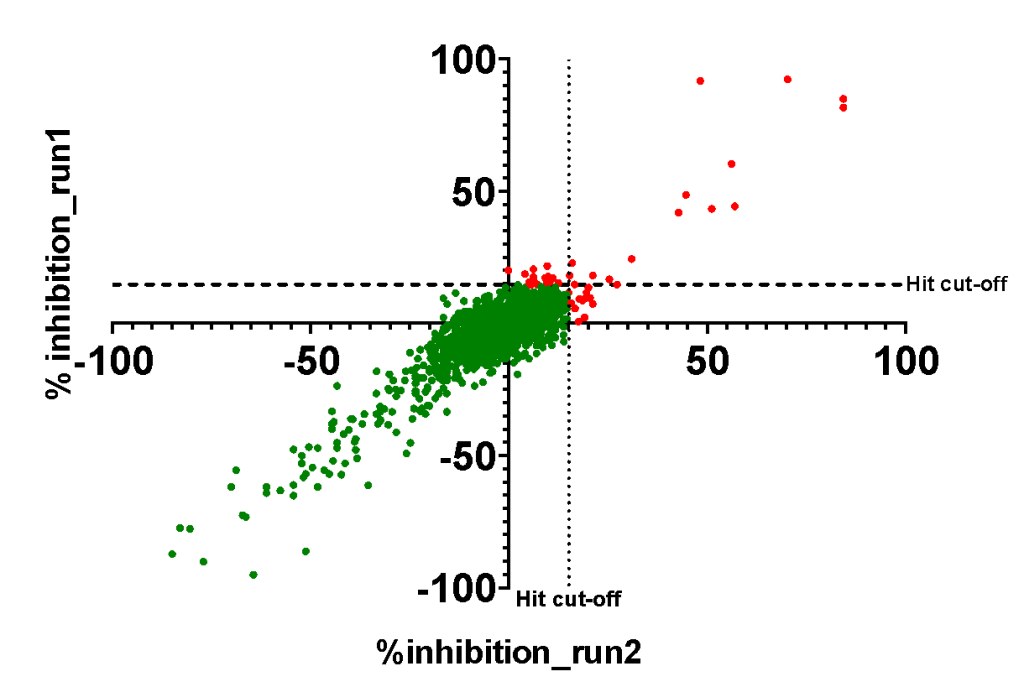
- 45 compounds with ≥15% inhibition (highlighted in red) in at least one of the two runs, in addition to one compound with unclear signal, were advanced to 20-point dose-response experiment by FP.
- All 46 compounds were tested for solubility and aggregation by dynamic light scattering (DLS)
- Eleven of the 46 compounds showed a measurable Kdisp < 150 µM in FP assay and their binding was tested in a 19F-NMR assay. Non-fluorinated compounds were tested with a Trp-fluorinated version of CBLB.
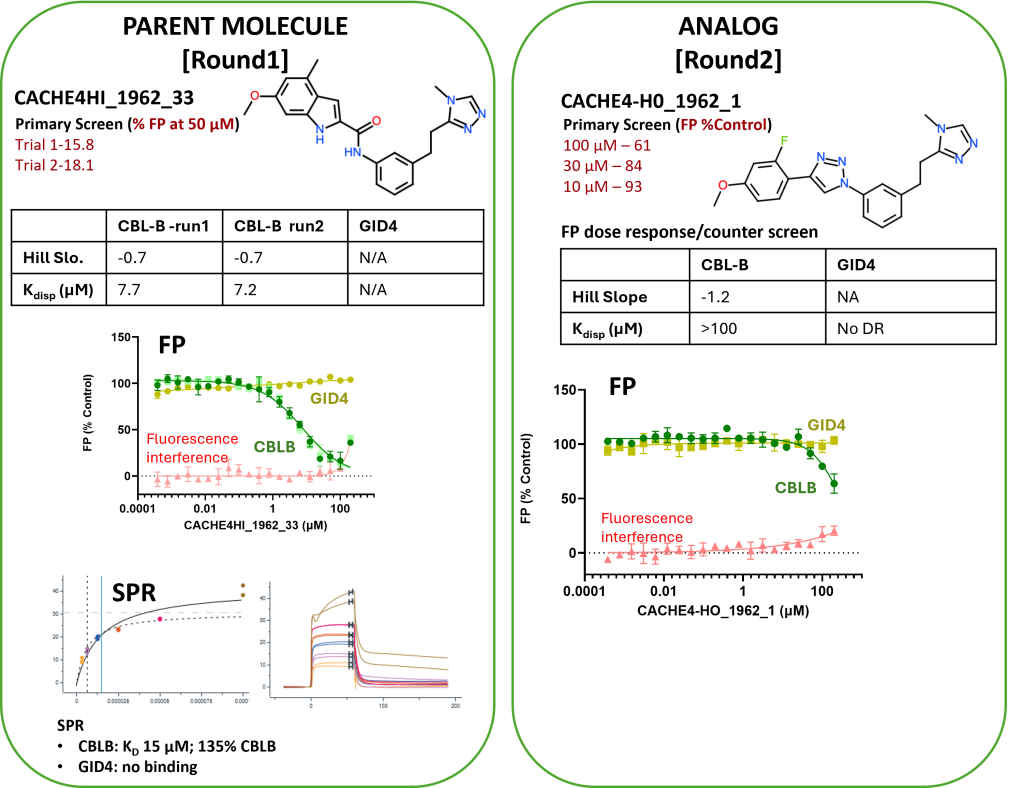
- Compounds CACHE4HI_1962_33 and CACHE4HI_1946_60 bound to CBLB in the NMR assay (details here).
- The eleven hit molecules from FP (details here ) were tested also by FP for binding to an unrelated protein, GID4.
- One compound with poor solubility and non-selective binding to GID4 was dismissed. Ten other compounds from nine participants were selective for CBLB and were advanced to Round 2: hit expansion.
ROUND 2 RESULTS
All Round 2 experimental data are provided here and here
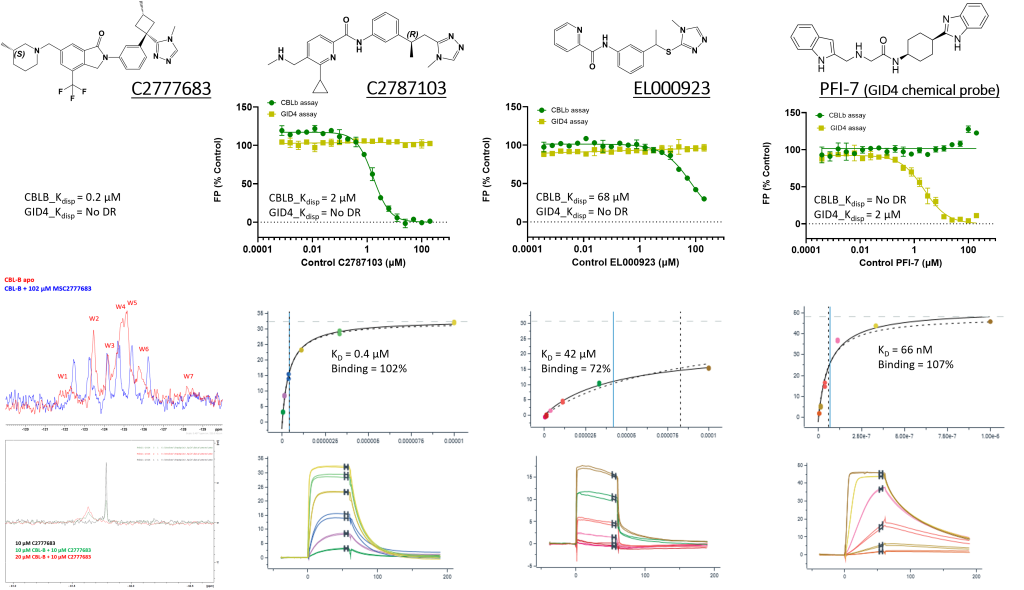
FP, 19F-NMR and SPR assays were validated with four positive controls:
- 201 analogs of Round 1 hits supplied by eight participants were advanced to Round 2 to confirm binding.
- The experimental pipeline was FP followed by NMR and SPR.
- All 201 compounds were screened at three concentrations (100, 30 and 10 µM) in duplicates in the FP assay.
- 22 compounds with at least ~20% inhibition (<80% FP Control/overflowed signal and no fluorescence interference) were further tested by FP and SPR with full dose-response against CBLB and the unrelated control protein GID4.
- In addition, 45 compounds that showed a dose dependent fluorescence interference (>120% FP Control at 100 µM) were advanced to dose-response step by SPR to avoid false negatives.
- The solubility of the 67 compounds was evaluated by DLS to determine the maximum concentration acceptable in the following dose response SPR assay.
- 19F NMR was used as an orthogonal binding assay to test 10 compounds that showed a dose dependent displacement of FP tracer and/or weak binding by SPR. Results are available here.
- Experimental results for all chemical series are compiled in these slides.
EVALUATION OF COMPUTATIONAL METHODS
- The biophysical data and SAR of Round 1 hits and their Round 2 follow-ups were evaluated by an independent Hit Evaluation Committee composed of industry experts in biophysics (Anders Gunnarsson, Astra Zeneca & Vera Puetter, Nuvisan), medicinal chemistry (Hartmut Beck, Bayer & Lars Wortmann, Boehringer Ingelheim), and computational chemistry (Pat Walters, Relay Therapeutics ) leading to a final score assigned to each Round 1 hit.
- In addition to the experimental binding data, the Committee evaluated any chemical liability and chemical novelty compared with previously disclosed compounds. The results are provided in Table 1
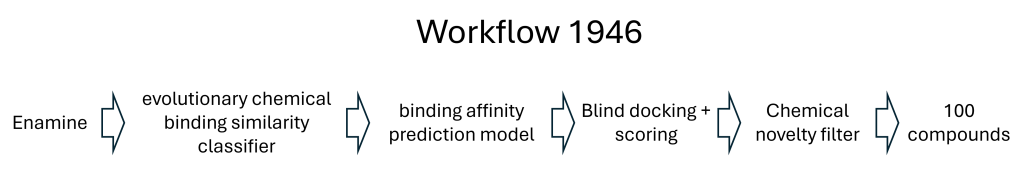
- Two Round 1 hits were clearly confirmed: CACHE4HI_1962_33 and CACHE4HI_1946_60. CACHE4HI_1962_33 had the most robust binding signal but is chemically related to compounds previously disclosed in the patent literature. CACHE4HI_1946_60 was chemically novel. A third compound, CACHE4HI_1946_25, was also binding CBLB, but with unclear specificity.
Schematic of the computational workflow leading to a chemically novel, experimentally confirmed CBLB ligand:
Scores from the CACHE hit evaluation committee:
| Rank | Workflow | Participant | Aggregated Score | Score of top chemically novel compound |
| 1 | 1946 | Keunwan Park, [ KIST] | 32.2 | 16.8 |
| 2 | 1962 | Wei Lu, Jixian Zhang [Galixir] | 21.4 | 0 |
| 3 | 1955 | 10.9 | 10.9 | |
| 4 | 1958 | 10.6 | 10.6 | |
| 5 | 1963 | 9.4 | 9.4 | |
| 6 | 1957 | 7.5 | 7.5 | |
| 7 | 1974 | 6.9 | 6.9 | |
| 8 | 2117 | 3.7 | 3.7 | |
| 9 | 1948 | 2.7 | 2.7 | |
| 10 | 1954 | 1.9 | 1.9 | |
| 11 | 1949 | 0 | 0 | |
| 12 | 1951 | 0 | 0 | |
| 13 | 1952 | 0 | 0 | |
| 14 | 1956 | 0 | 0 | |
| 15 | 1960 | 0 | 0 | |
| 16 | 1961 | 0 | 0 | |
| 17 | 1964 | 0 | 0 | |
| 18 | 1965 | 0 | 0 | |
| 19 | 1966 | 0 | 0 | |
| 20 | 1969 | 0 | 0 | |
| 21 | 1972 | 0 | 0 | |
| 22 | 1973 | 0 | 0 | |
| 23 | 1975 | 0 | 0 |
In a separate evaluation scheme, all 1688 compounds selected in Round 1 were merged and participants were asked to predict active molecules from this merged library. This is a complementary exercise, as here all participants screen the same set of compounds. But since only three compounds were confirmed as active, only two with clear specificity, this evaluation step was deemed not statistically significant and dismissed.
Top Confirmed Compounds: